Introduction
Biomedical research emphasizes the reproducibility of scientific research by independent researchers, as the results have direct effects on human health. The increasing complexity of modern research, along with inadequate documentation, equipment and resource limitations, and environmental variability, has contributed to a “reproducibility crisis” [1]Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016). [2] Ioannidis JPA (2014) How to Make More Published Research True. PLoS Med 11(10): e1001747.[3] Samuel S, König-Ries B. Understanding experiments and research practices for reproducibility: an exploratory study. PeerJ. 2021 Apr 21;9:e11140. doi: 10.7717/peerj.11140.. NexusGraph offers an advanced Natural Language Processing (NLP) solution to this crisis. It synthesizes a comprehensive, accessible inventory of reagents, equipment, and other materials to allow for standardization to replicate studies. Furthermore, NexusGraph extracts additional elements, including author and material suppliers, along with synthesizing instructional guidance to increase reproducibility.
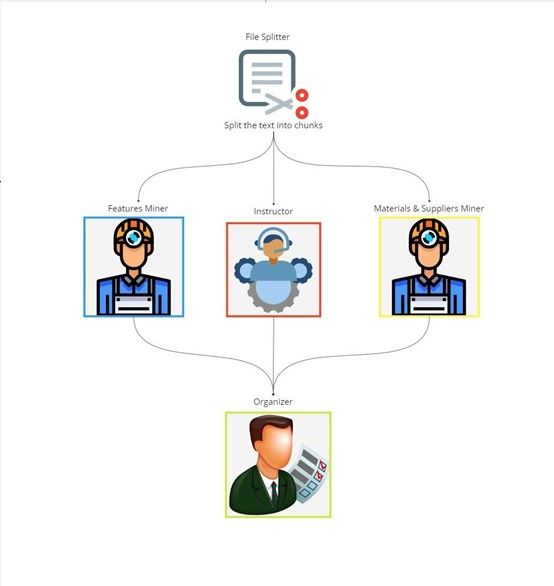
Figure 1. NexusGraph worker element pipeline starting with input file splitting, followed by feature mining, instructor, and materias and supplier mining, output by the organizer into a relational database
Description of our System
NexusGraph extracts, analyzes, and outputs information from scientific papers through a 5-component pipeline. The 5 stepwise components being:
- File Splitter
- Feature Miner
- Materials and Supplies Miner
- Instructor
- Organizer
NexusGraph initiates the extraction of data by passing the inputted paper through the File Splitter. This File Splitter divides the text through a set a predefined keywords (e.g., 'TITLE', 'ABSTRACT', 'METHODS') for a granular analysis of the document. The design ensures that each section precedes the corresponding keyword to precisely section appropriate sections. The Feature Miner utilizes the ChatGPT API to extract demographic information, including the paper's title, authors, affiliations, and category. This miner is optimized to balance accuracy and creativity, to identify unique titles and categories for the metadata output. The Materials and Supplies Miner further utilizes the ChatGPT API to extract specific materials, suppliers, and experiments. The Materials and Supplies Miner configures the ChatGPT API to a higher degree of temperature. This enables it to infer and add elements not explicitly mentioned. The instructor creates a step-by-step protocol for the methodology that guides the researcher to replicate the experiment. The instructor works at the highest degree of temperature to allow for creative, innovative, and practical guidance for the researcher. The Organizer merges the outputs from the Feature Miner, Materials and Supplies Miner, and Instructor into one JSON file that is inserted into a relational database. This allows for structured and efficient retrieval of comprehensive records or specific details.
Degree of Deployment
NexusGraph has been deployed and used to analyze a set of 50 academic papers in material science. It has successfully extracted 163 authors, 50 titles, 473 materials, and 70 suppliers. It has also generated 50 unique sets of instructions for all 50 papers. A comparative analysis between manual processing on a single paper showed successful identification of title, author details, and tags - metadata. Materials, suppliers, and applications also showed high precision. However, the Instructor experience difficulties and diverged from the manual processing. NexusGraph has many similarities to manual processing, but challenges are present for future enhancement.
References
- Baker, M. 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016). https://doi.org/10.1038/533452a
- Ioannidis JPA (2014) How to Make More Published Research True. PLoS Med 11(10): e1001747. https://doi.org/10.1371/journal.pmed.1001747
- Samuel S, König-Ries B. Understanding experiments and research practices for reproducibility: an exploratory study. PeerJ. 2021 Apr 21;9:e11140. doi: 10.7717/peerj.11140. PMID: 33976964; PMCID: PMC8067906.



